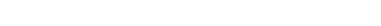Cell Image Analysis
バイオイメージング技術は急速に発展してきており,様々な生命現象が新たに観察できるようになり,生物学分野に革新を起こしつつあります.このような研究において,より生体内に近い状態での解析は非常に重要になります.しかし,これまでのディッシュ上での観察と比べ,生体内では,細胞や分子等の観察対象が超高密度・大量に分布しており,画像データからの自動定量評価が難しいという課題があり,生物学分野における定量化研究の障壁となっています.静的・動的データの定量的な評価を行うためには,個々の細胞のセグメンテーション及びトラッキングをすることが求められます.しかし,バイオイメージ画像においては,人や車等の一般物体の追跡と比べ,「低コントラストで境界が非常に曖昧」「形状が様々に変化する」「対象同士の見た目が非常に類似している」といった様々な課題があります.このような課題を解決し,自動定量化を実現するため,自動細胞領域検出及びトラッキング等の細胞画像認識手法の研究開発を行っています.
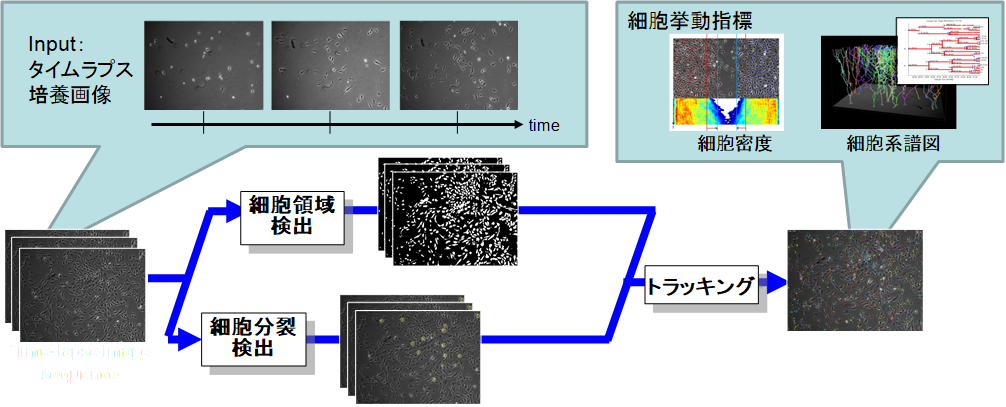
Medical Image Analysis
医療の質・安全性の向上、高度化、効率化、均てん化のため、ICTを活用して医療画像に代表される医療ビッグデータを収集・利活用するための基盤が求められています。本研究開発では、大量の医療画像データを対象とした深層学習等、最新の画像解析技術を利用した新しい技術開発を行っています。単純に課題設定がされた公開データセットを利用した研究開発ではなく、医師と密接に連携し、どのような課題に取り組むべきか、どのように学習データを作成すればよいかを含めて議論し,研究を進めています。特に、病理画像や内視鏡画像を対象とした研究開発を行っています。
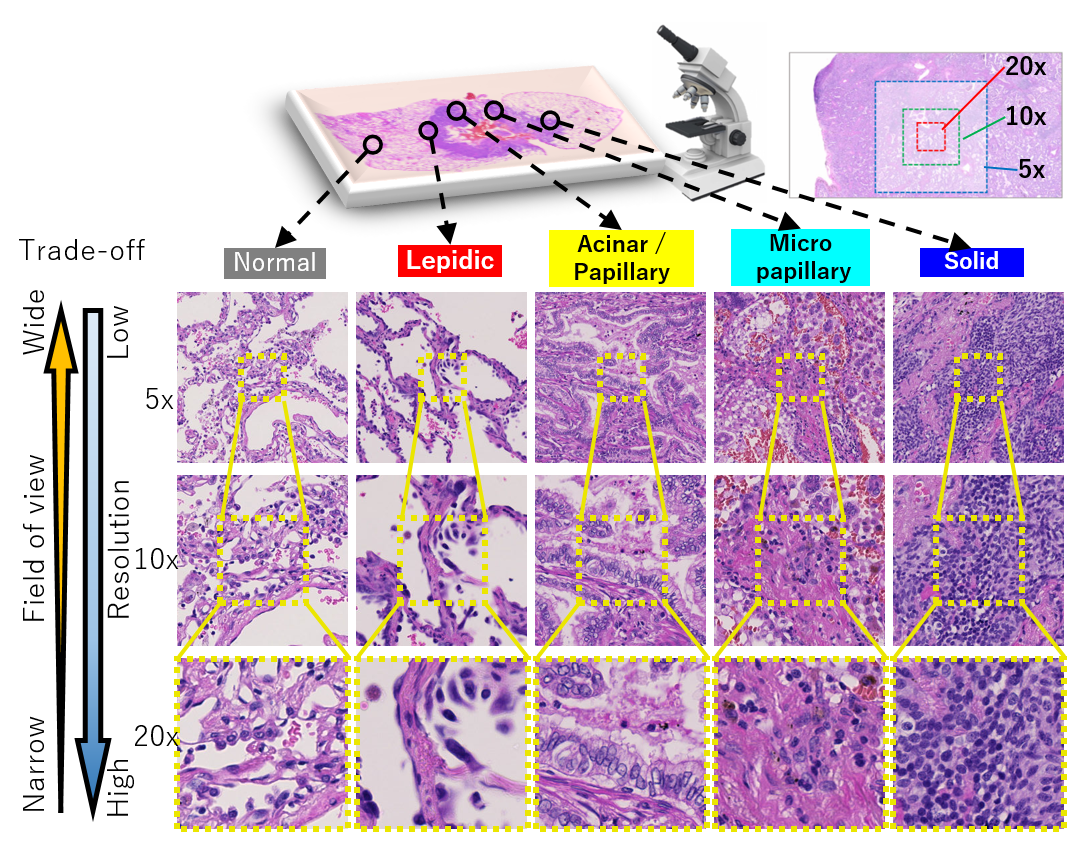
光超音波イメージングと血管解析
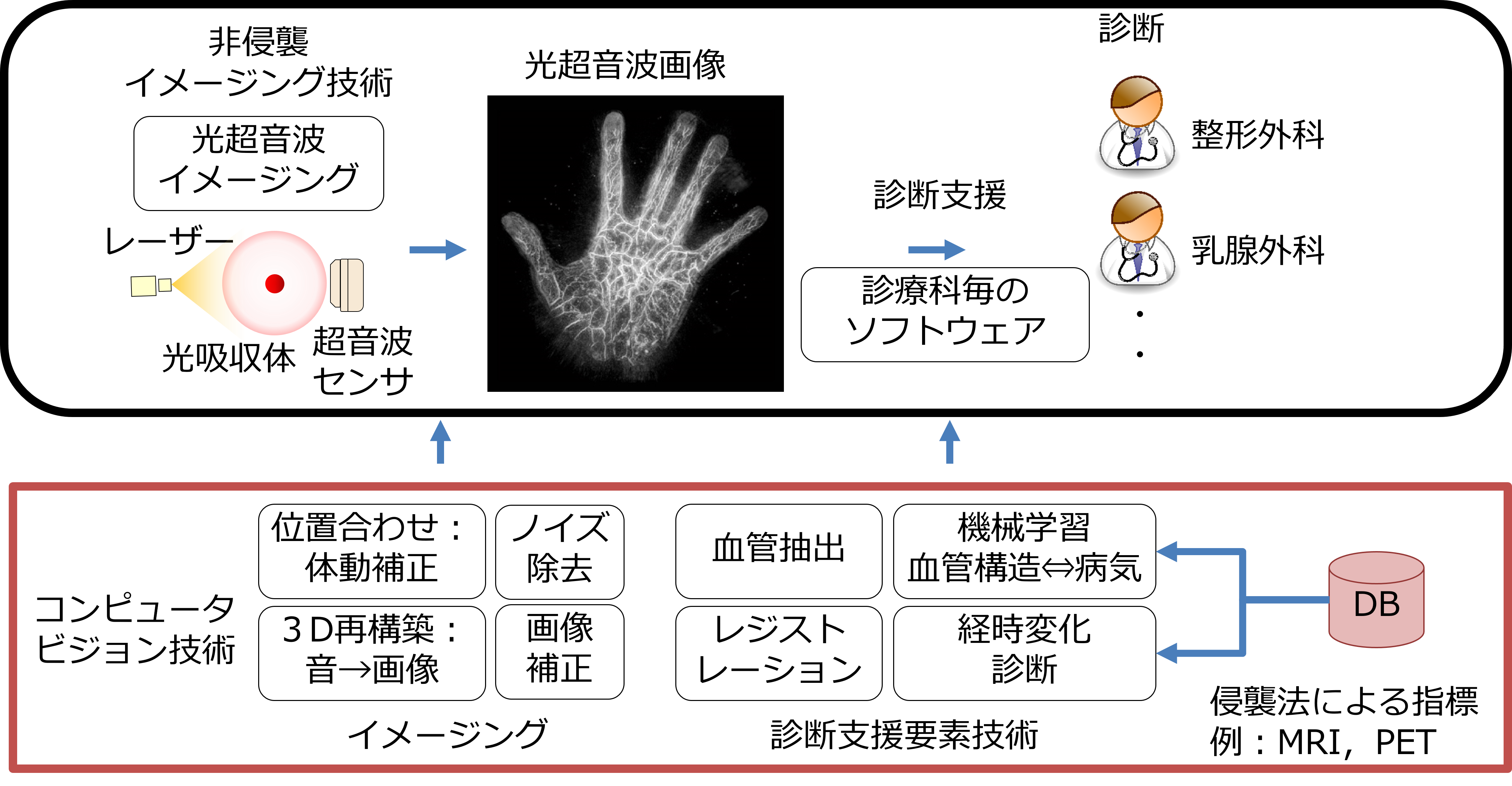
高齢化社会の到来に伴い、健康長寿で豊かな生活を実現し、病気や介護への不安を解消させる技術サポートが求められています。九大は、病気の早期診断や超精密検査の実現を目指すImPACTに参加し、生体や物体内部を非侵襲・非破壊でリアルタイム三次元可視化する光超音波イメージングの高度化を行いました。光超音波システムは、レーザー照射により発生する超音波を検出し可視化する最先端計測技術です。この技術は、非侵襲・非破壊である上に、透過して深部まで照射できる光と超音波の両方の特性を活かし、肉眼では見えない様々な対象の可視化を可能にします。本研究では、コンピュータビジョン技術により、鮮明な画像を得るイメージング技術の高度化や、様々な情報を用いた画像解析による診断支援を行っています。例えば、撮影中の患者の体動による画質劣化に対して、画像の位置合わせにより患者の動きを補正し、画質改善した診断しやすい画像を提供できるようになります。また、疾病に関係が深い血管状態を把握するため、血管構造の自動抽出技術の開発を進めています。