Medical Image Analysis
In order to improve the quality and safety of medical care, sophistication, efficiency, and equalization, a foundation for collecting and using medical big data represented by medical images using ICT is required. In this research, we are developing new technologies using the latest image analysis technology, such as deep learning for large amounts of medical image data. In addition to research and development that uses public datasets simply set up with tasks, we will collaborate with doctors and discuss what tasks we should work on, including how to create training data. Current our main targets are pathological images and endoscopic images.
Adaptive Weighting Multi-Field-of-View CNN for Semantic Segmentation in Pathology
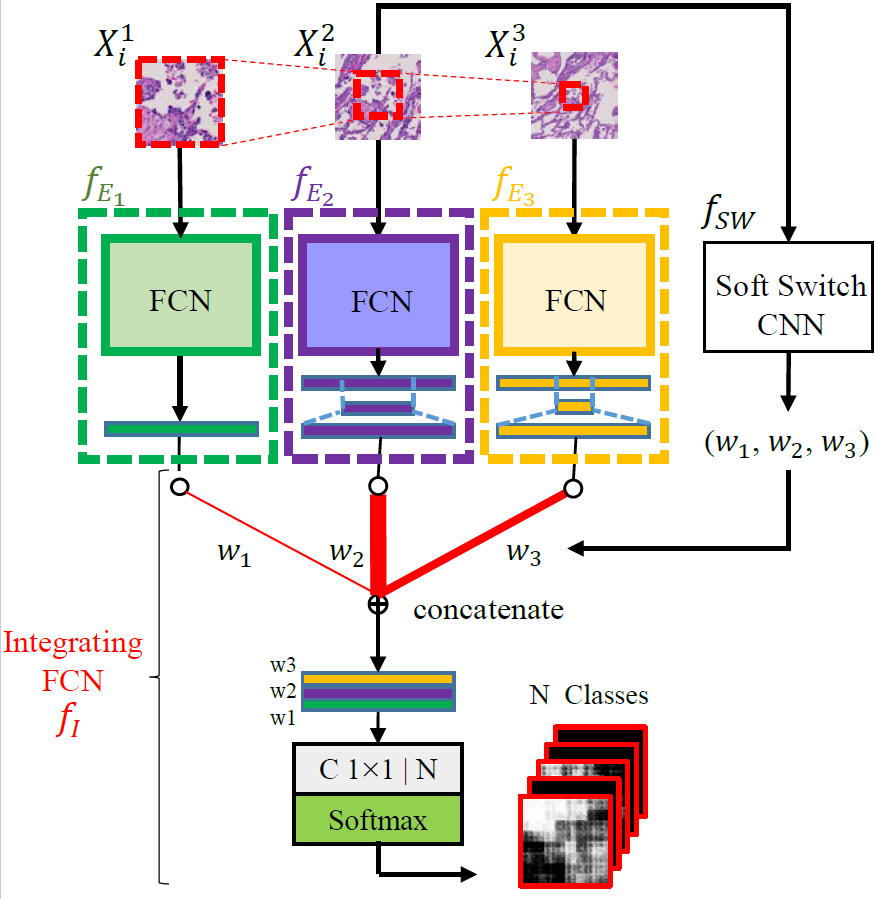
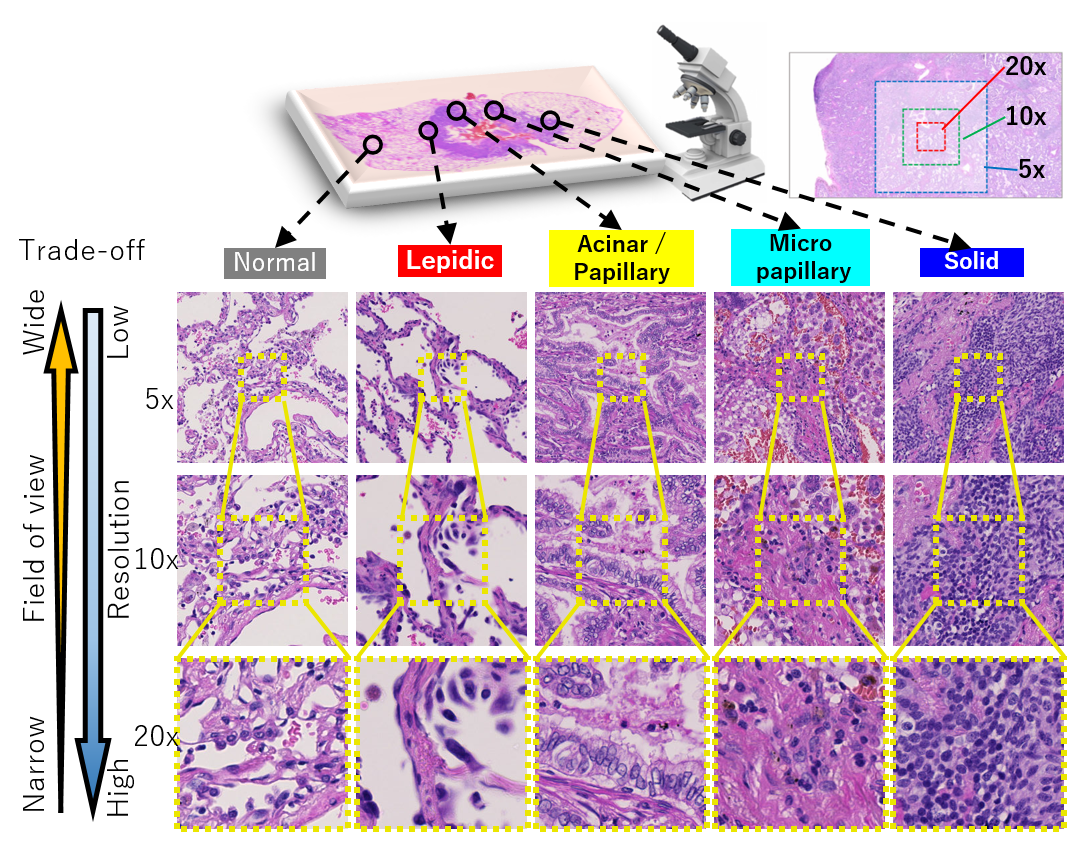
Automated digital histopathology image segmentation is an important task to help pathologists diagnose tumors and cancer subtypes. For pathological diagnosis of cancer subtypes, pathologists usually change the magnification of whole-slide images (WSI) viewers. A key assumption is that the importance of the magnifications depends on the characteristics of the input image, such as cancer subtypes. In this paper, we propose a novel semantic segmentation method, called Adaptive-Weighting-Multi-Field-of-View-CNN (AWMF-CNN), that can adaptively use image features from images with different magnifications to segment multiple cancer subtype regions in the input image. The proposed method aggregates several expert CNNs for images of different magnifications by adaptively changing the weight of each expert depending on the input image. It leverages information in the images with different magnifications that might be useful for identifying the subtypes. It outperformed other state-of-the-art methods in experiments.
Endoscopic Image Clustering with Temporal Ordering Information Based on Dynamic Programming
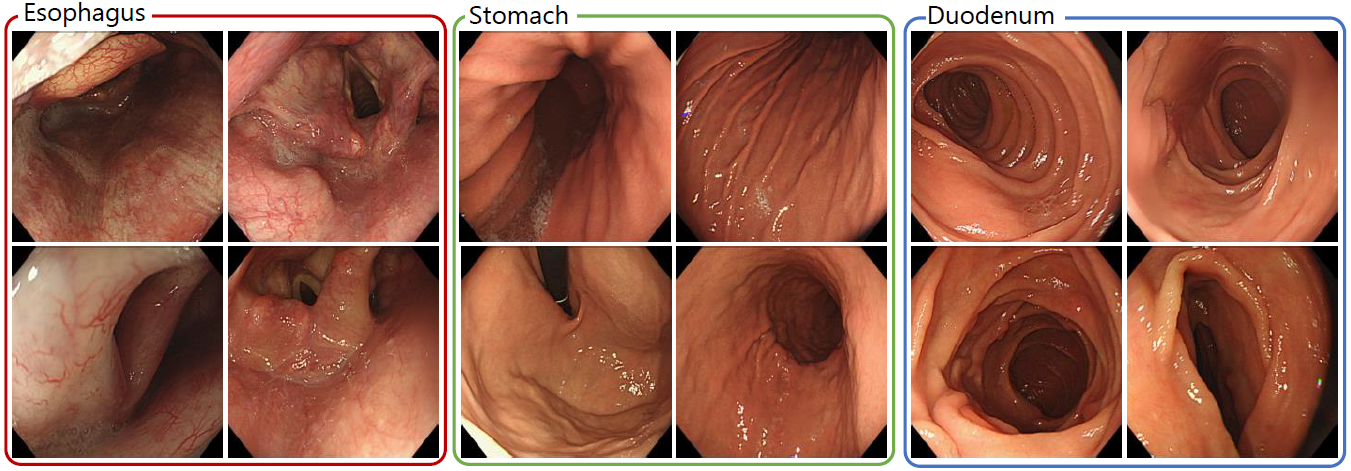
In this paper, we propose a clustering method with temporal ordering information for endoscopic image sequences. It is difficult to collect a sufficient amount of endoscopic image datasets to train machine learning techniques by manual labeling. The clustering of endoscopic images leads to groupbased labeling, which is useful for reducing the cost of dataset construction. Therefore, in this paper, we propose a clustering method where the property of endoscopic image sequences is fully utilized. For the proposed method, a deep neural network was used to extract features from endoscopic images, and clustering with temporal ordering information was solved by dynamic programming. In the experiments, we clustered the esophagogastroduodenoscopy images. From the results, we confirmed that the performance was improved by using the sequential property.
- [S. Harada, H. Hayashi, R. Bise, K. Tanaka, Q. Meng, and S. Uchida, IEEE EMBC,2019, accepted]
Scribbles for Metric Learning in Histological Image Segmentation
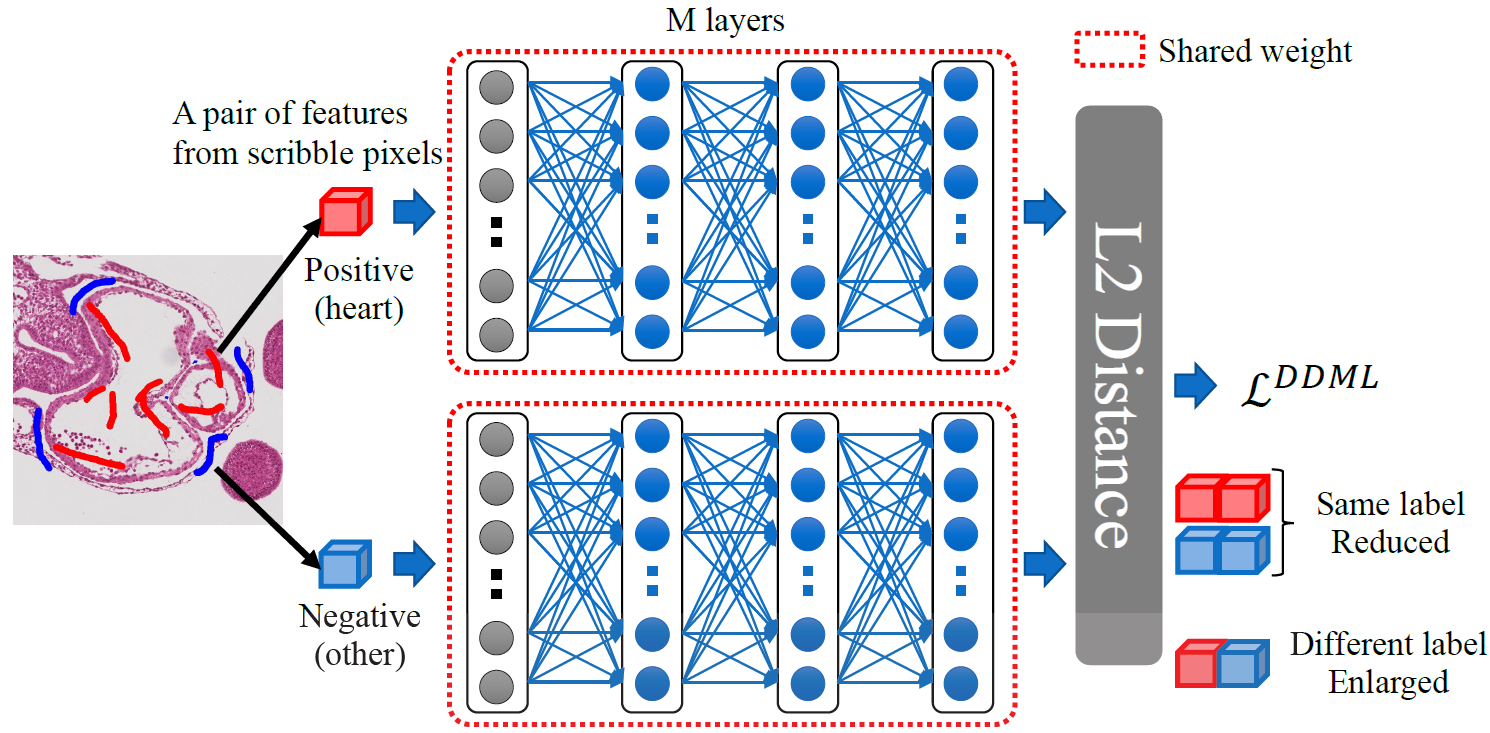
Segmentation is a fundamental process in biomedical image analysis that enables various types of analysis. Segmenting organs in histological microscopy images is problematic because the boundaries between regions are ambiguous, the images have various appearances, and the amount of training data is limited. To address these difficulties, supervised learning methods (e.g., convolutional neural networking (CNN)) are insufficient to predict regions accurately because they usually require a large amount of training data to learn the various appearances. In this paper, we propose a semi-automatic segmentation method that effectively uses scribble annotations for metric learning. Deep discriminative metric learning re-trains the representation of the feature space so that the distances between the samples with the same class labels are reduced, while those between ones with different class labels are enlarged. It makes pixel classification easy. Evaluation of the proposed method in a heart region segmentation task demonstrated that it performed better than three other methods.
- [D. Harada, R. Bise, H. Tokunaga, W. Ohyama, S. Oka, T. Fujimori, and S. Uchida, IEEE EMBC,2019, accepted]
